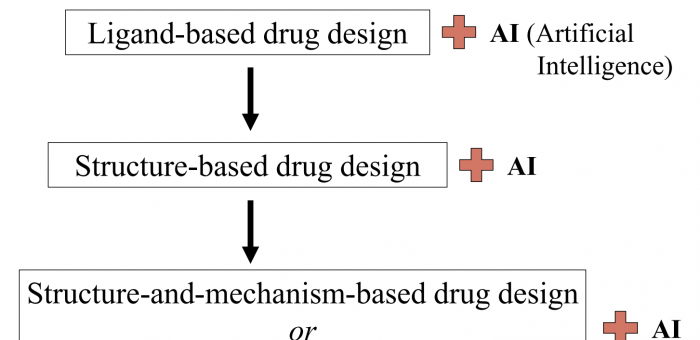
Nick Teets
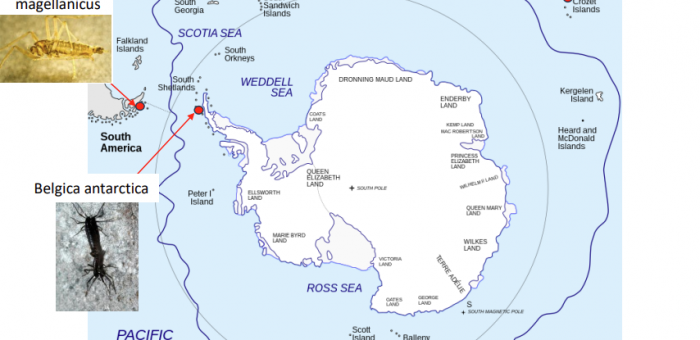
Website: https://teetslab.com/Scholars@UK: https://scholars.uky.edu/en/persons/nicholas-teetsSlides from CCS/RCD Seminar on 3/19/2024: 20240319-Teets-Seminar.pdf Genomics and transcriptomics of extreme insects Insects are the most diverse and abundant animals on the planet, but Antarctica is the exception to that rule. Only a handful of species can survive Antarctica’s harsh terrestrial environments, and these insects have unique adaptations for coping with extreme abiotic conditions and long winters. Our lab is interested in the physiological and molecular mechanisms by which these insects survive (and thrive) at the bottom of the earth. Antarctic insects are difficult to collect and currently unamenable to laboratory rearing, so genomics provides a powerful tool for identifying mechanisms that underpin adaptation to extreme environments. We use a combination of transcriptomics, whole genome sequencing, and population genomics to characterize the molecular and evolutionary genetic processes…